Yi-C.- W., Yang, C.-H., Hung J.-H
【Motivation】
Genetic engineering is an indispensable field forbiomedical researches nowadays. Next generation sequencing (NGS ) enables high-throughput sequencing, in which short DNA fragments can be
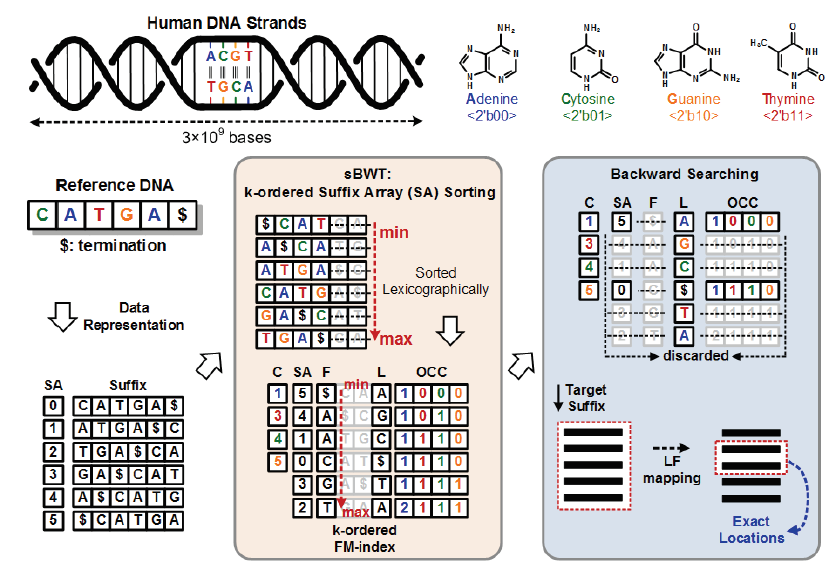
sequenced in a massively parallel fashion. However, the essential algorithm behind the succeeding NGS data analysis, DNA mapping, is still excessively time consuming. Based on the memory-efficient sBWT algorithm, this work is the first integrated NGS data processor for the practical NGS data analysis. The k-ordered FM-index used in the sBWT algorithm is leveraged to improve torage capacity and reduce hardware complexity. The proposed NGS data processor realizes the sBWT algorithm through novel Bucket Sorting, Suffix Grouping, and Suffix Sorting circuits. The most complicated part, SA sorting, can now be finished within ten minutes.
【System model and breakthrough】
The conventional algorithms uses hash table to perform DNA mapping, but too much memory space is required. The FM-index based induces large memory footprints and thus is not a proper way for hardware implementation. In this work, sBWT algorithm is utilized to construct the k-ordered FM-index, which can greatly reduce the hardware complexity. In addition, it can also scale down the memory space to meet the system requirement and bring a feasible solution for hardware realization within 1 Gigabyte.The software implementation introduces heavy computation loads for sorting. In this work, massively parallel insertion sorting elements are utilized to reduce the sorting latency. By exploiting bucket sort with up/down sampling scheme, the computation complexity
and workload are greatly reduced, thus enhancing the throughput for FM-index construction.Conventional software algorithm is not able to execute in parallel when performing target string matching. In this work, several parallel matching modules are used to recover the down-sampled data in FM-index. Compared to the baseline design, the searching latency is saved by 98%. The key design parameters are analyzed and selected dynamically to reach the optimal performances for different lengths of DNA sequences. Parameters are adaptively derived to achieve maximal earching throughput under given memory spaces and DNA sequence.
【Experimental Results and Conclusion】
Fabricated in the 40nm process, the chip integrates 7.4M gates in 7.84mm2 area. The power dissipation is 135mW at 200MHz from a 0.9V supply voltage. This work is the first dedicated NGS data processor that implements both SA sorting and backward searching on silicon. It achieves more than 8,000x higher energy efficiency to high-end GPU solutions.